News
Congratulations to Chava for successfully defending his doctoral dissertation entitled: “Monitoring the spread of plasmid-mediated antibiotic resistance in agricultural soil.”
The first two chapters from his work are available online! Chapter 1: Plasmids, a molecular cornerstone of antimicrobial resistance in the one health era. In this review, we outline the global significance of antimicrobial resistance with a focus on plasmids as key players in the dissemination of antibiotic resistance genes. Utilizing a One Health approach, we explore the interconnectedness of habitats and discuss factors that influence the ecology and evolution of plasmids.
Chapter 2: Detection of rare plasmid hosts using a targeted Hi-C approach. We first established the detection limit of Hi-C, a proximity ligation method, for associating a plasmid with its host in soil. For a plasmid to be associated with its host, the pair needed to be present at, or above, a relative abundance of .001%. Then, we combined Hi-C with a target capture approach aimed at enriching for DNA of the focal plasmid. This enrichment approach, referred to as Hi-C+, increased the detection limit of Hi-C 100-fold.
Congrats to Chava on recently receiving the 2022 Paul Joyce Memorial Award!
Eva was named an American Association for the Advancement of Science Fellow in recognition of her pioneering work on the ecology and evolution of drug-resistant bacteria. Congrats Eva! https://www.iids.uidaho.edu/news.php?newsid=112
Congrats to former undergraduate student Aaron Law for being first author on this paper showing that biosolids used as fertilizer can still spread drug resistance plasmids to pathogens! https://www.frontiersin.org/articles/10.3389/fmicb.2021.606409/full
Better late than never: former postdoc Wesley Loftie-Eaton persevered and got this cool paper published many years after he left the lab. The findings suggests that rare gut microbiome members should not be ignored as potential reservoirs of multidrug resistance plasmids from food. See https://journals.asm.org/doi/10.1128/AEM.02735-20
Congratulations to Dr. Thibault Stalder and Dr. Erin Mack for receiving the Outstanding Team Award as Team Leader and Team Member, respectively, together with Cindi Brinkman from Dr. Coat’s lab in Civil Engineering. During the past couple of months, they have been working in collaboration with other individuals at the University of Idaho to sample wastewater for SARS-Cov-2.
Congrats to Clint on recently receiving the 2021 Paul Joyce Memorial Award!
Researchers at the University of Idaho, including our very own Dr. Thibault Stalder, have partnered with the City of Moscow to improve wastewater testing for the presence of SARS-CoV-2! The team hopes to develop an early warning system for spikes in local cases. Read more about it through the University website and through a local paper.
Congrats to Dr. Hannah Jordt and team for her paper in Nature Ecology & Evolution on the emergence of multi-drug resistance!
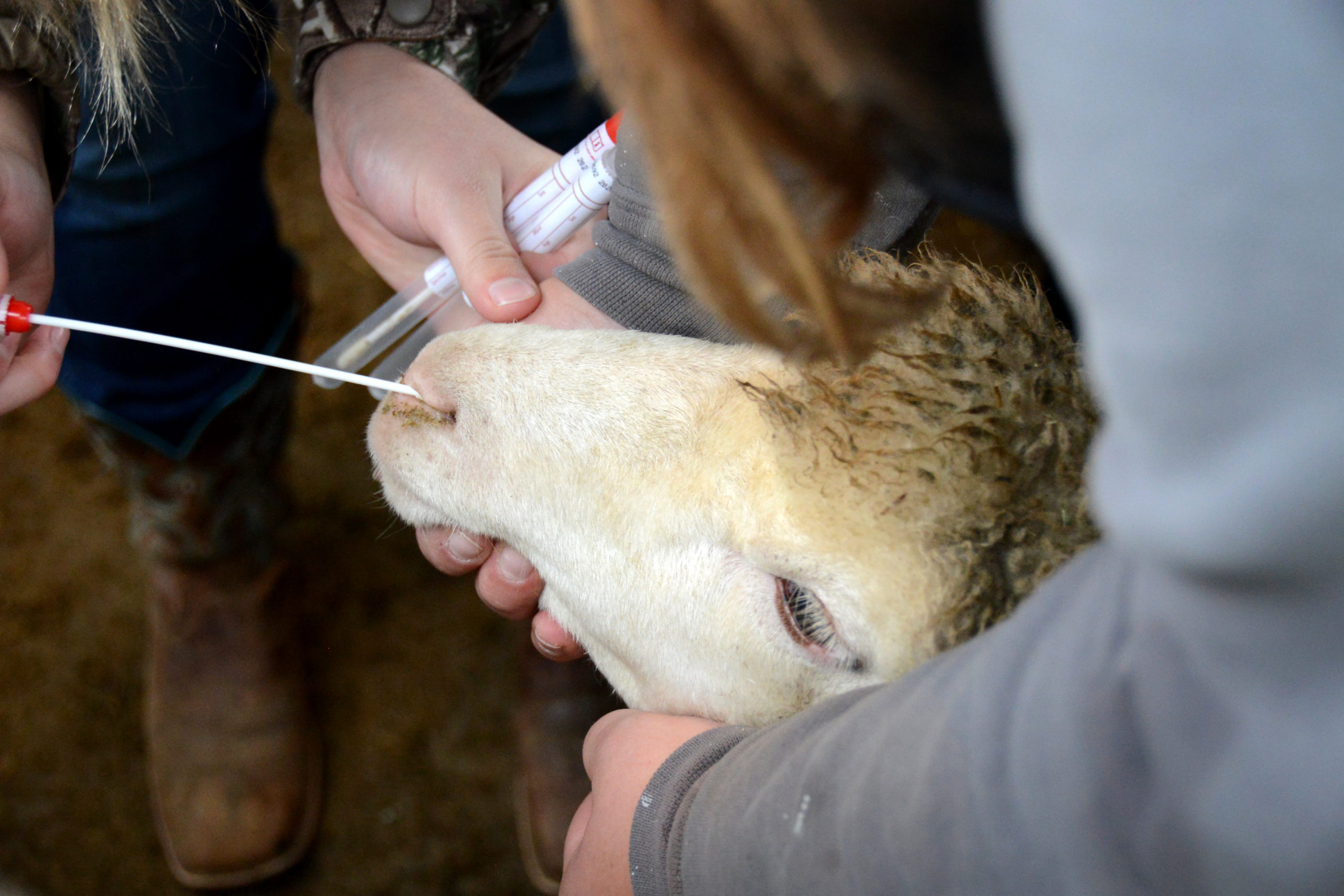
Thibault Stalder is currently working on a project that involves the collection of nasal swabs from young sheep. The goal of the project is to aid in the creation of a vaccine to protect sheep against the Mycoplasma ovipneumoniae (Movi) bacteria. Read more about it on the IBEST website!
Congrats to Dr. Thibault Stalder and team for his paper in MBE on the evolution of plasmid persistence in biofilms!

Congrats to Clinton Elg for advancing to candidacy in the Bioinformatics and Computational Biology PhD program!
This past summer was full of traveling for our lab as our lab members attended three different conferences to share updates on their latest research projects. Our lab members attended the Gordon research conference on Microbial Population Biology (Eva, Thibault, and Clint), Multi-omics for Microbiomes Conference (Thibault and Chava), and
Thibault: Usage of proximity-ligation (Hi-C) to determine hosts of anitbiotic resistance genes and mobile genetic elements in microbiomes.
Clint: Compensatory deletion of putative methylation gene increases plasmid persistence in absence of selective pressure.
Chava: Usage of the Hi-C method to detect plasmid-host associations in soil.
Congrats to Thibault Stalder for his recently accepted paper in ISME J on using Hi-C to link plasmids and antibiotic resistance genes to their bacterial hosts in a wastewater microbiome! His paper was also recently featured on an article in BioSpace!
Congrats to Clinton Elg for obtaining a prestigious NSF graduate research fellowship!
Congrats to undergraduate researchers Aaron Law and Sue Winger for graduating and winning the Undergraduate Research Award and one of the dean’s awards in the College of Science!